Deoxyribonucleic acid or DNA is rightly referred to as the molecule which accounts for ‘Unity in Diversity’ in living organisms and this molecule has provided valuable biological insights including human and animal health, biodiversity, agriculture, forensics, evolution, and much more. The list of applications continues to expand with the availability of DNA or RNA sequences of different organisms through new generation sequencing technologies.
So, the biological information present in the order of nucleotides of the genetic material or ‘genome’ of an organism includes the presence of different genes and proteins and their interaction for the overall functioning of an organism. But, the complete biological information about an organism comes not only from its ‘genome’ but from its ‘microbiome’ as well. Yes, microbes are part of every organism and every organism is part of them including humans. In fact, the density of microbial cells in a human gut amounts to ~ 1012 cells/ml which can be stated as an additional organ, the genome of which exceeds the size and metabolic function of the host itself. The human microbiota consists of around 150 times more genes than are found in the entire human genome (Goodrich et al. 2017).
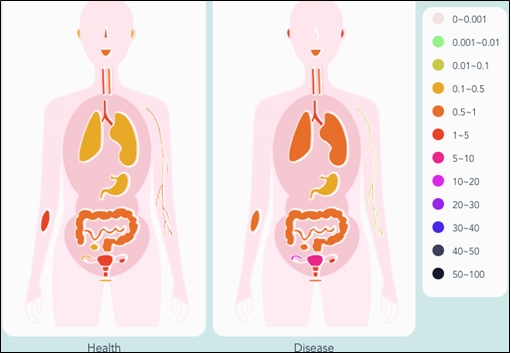
The microbes are part of human existence right from the time of birth till death and the microbiome can change significantly in various stages of life. It can be altered by various external factors including diet, exercise, age, disease, use of drugs, environment, etc. Also, the composition of microbial community is different and unique for different individuals. In fact, the gut microbiome in healthy adults can be stable for years (Faith et al. 2013). Thus, each individual has a specific ‘microbial fingerprint’ which is different from other individuals. Such ‘microbial signatures’ can provide useful information about a particular disease or may also provide cues for its treatment. These include ‘marker taxa’ which are relatively different in abundance at various body sites or between healthy and diseased state of an individual both at the species and genus levels. For instance, bacterial signatures were studied in patients with metabolic disorders like inflammatory bowel disease (IBD). It was found that actinobacterial genera including Actinomyces, Bifidobacterium and Eggerthella were present in higher proportion in IBD and metabolic disease (MD) patients while five genera Alistipes, Eubacterium, Roseburia, Feacalibacterium and Akkermanisa were decreased in abundance in both IBD and MD patients. Since these genera are known producers of short chain fatty acids (SCFAs) which are beneficial to health, thus, it’s obvious that their decreased number can cause poor health (Proffitt et al. 2020). Also, the microbial signatures associated with oral health and periodontitis include bacterial species like Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola and Aggregatibacter actinomycetemcomitans (Pozhitkov et al. 2015). Recently, researchers have identified the microbial signatures associated with posttreatment Lyme disease syndrome (PTLDS), which is characterized by an increase in Blautia, and a decrease in Bacteroides (Morrissette et al. 2020). Further, in order to facilitate the identification of site- and/or disease-specific marker microbes, Jin et al. 2021 have developed a curated database called mBodyMap. This database can be explored to find out the distinct microbial profiles across the human body sites and their associations with health and diseases. For example, the relative abundance of genus Megasphaera in nose is ~3.01% in healthy individuals while it is almost completely absent in diseased state (Figure 1). On the other hand, its relative abundance increases significantly from 1.76% (in health state) to 5.45% in uterus in diseased state.
Such information can prove immensely useful in development of diagnostic and therapeutic tools in which health-related microbial community can be transplanted to the diseased patient. For instance, fecal microbiota transplantation has been used successfully to treat recurrent Clostridium difficile infection (rCDI). Also, oral microbiota transplant holds therapeutic promise for common diseases like dental caries and periodontitis. Thus, with the availability of microbiome data and analysis, cost-effective and accessible strategies will be available to general public in near future. Having said that, it should be noted that use of human microbial signatures cannot replace the conventional genome-based approaches to treat human diseases or disorders but can augment those approaches for effective treatment of diseases. To conclude, microbes are an inseparable part of humans and human microbiome studies have the potential to solve many problems related to human health and disease.
References
- Goodrich, Julia K.; Davenport, Emily R.; Clark, Andrew G.; Ley, Ruth E. (2017). The Relationship Between the Human Genome and Microbiome Comes into View. Annual Review of Genetics, 51(1).
- Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013;341:1237439.
- Proffitt, C., Bidkhori, G., Moyes, D., & Shoaie, S. (2020). Disease, Drugs and Dysbiosis: Understanding Microbial Signatures in Metabolic Disease and Medical Interventions. In Microorganisms (Vol. 8, Issue 9, p. 1381). MDPI AG.
- Hanbo Jin, Guoru Hu, Chuqing Sun, Yiqian Duan, Zhenmo Zhang, Zhi Liu, Xing-Ming Zhao, Wei-Hua Chen, mBodyMap: a curated database for microbes across human body and their associations with health and diseases, Nucleic Acids Research, 2021.
- Morrissette, M., Pitt, N., González, A., Strandwitz, P., Caboni, M., Rebman, A. W., Knight, R., D’Onofrio, A., Aucott, J. N., Soloski, M. J., & Lewis, K. (2020). A Distinct Microbiome Signature in Posttreatment Lyme Disease Patients. In M. S. Gilmore (Ed.), mBio (Vol. 11, Issue 5). American Society for Microbiology.