A novel corona virus, SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) emerges as a pandemic disease with high mortality than the episodes caused by SARS-CoV (Severe acute respiratory syndrome coronavirus) and MERS-CoV (Middle East respiratory syndrome corona virus) in the last 20 years (1). The entry of SARS-COV-2 into the host cell is mediated by ACE2 (Angiotensin-converting enzyme 2) receptors which are mainly involved in regulation of blood pressure, fluid balance and inflammation (2). After primary infection, SARS-CoV-2 grows at a faster rate and spread to various organs especially lungs (3, 4). The host cell orchestras a wide range of immune responses to counteract the viral invasion.
The innate and adaptive immunity synergise together for initiating the immune responses such as production of proinflammatory cytokines, recruitment and activation of lymphocytes for controlling the viral replication, inflammation, limiting the spread of the virus, removal of infected cells and development of antigenic memory (5). Tissue injury caused by the virus could induce the exaggerated production of proinflammatory cytokines, the recruitment of proinflammatory macrophages and granulocytes (6). This results in the cytokine storm progressing to ARDS (Acute respiratory distress syndrome). The level of the proinflammatory cytokines and subsets of immune cells is correlated to the severity of SARS-CoV-2 infection.
STRUCTURAL FEATURES OF SARS-COV-2
Corona viruses are enveloped, positive sense single stranded, large RNA genome of 26 to 32 kb with distinctive club-like spikes projecting from their surface. They belong to the Coronaviridae family and are classified into four genera: α, β, γ, and δ based on the genome structure and phylogenetic analysis. The corona viruses of α and β genera typically infect human and mammals, whereas the coronaviruses of the γ and δ genera specifically infect birds (7). The genome structure of SARS-CoV-2 is similar to other typical coronaviruses that the majority of the viral RNA genome is translated into viral replicase transcriptase complex and the remaining minor portion is translated into structural and other accessory proteins. The nucleocapsid of the virion composed of the RNA genome and nucleocapsid protein (N) covered by the spike glycoprotein (S), membrane protein (M) and the envelope protein (E) (8). The genome structure of SARS-CoV-2 is similar to other typical corona viruses that the majority of the viral RNA genome is translated into viral replicase transcriptase complex and the remaining minor portion is translated into structural and other accessory proteins. S protein in its large ectodomain contains a receptor binding S1 subunit and a membrane fusing S2 subunit (2, 9). The spike protein of SARS-CoV-2 contains a multibasic S1/S2 site (10). For successful entry of virus into the cell, the S1/S2 site must be cleaved by the host cell proteases present on the cell membrane to enable exposure of the fusion sequence and permitting the fusion of host and viral membrane. The genomic sequencing of the β SARS-CoV isolated from humans was found to be identical with that of the sequence derived from bats and pangolins (11).
ACE2 RECEPTOR, FUNCTION AND ITS ROLE IN INFLAMMATION
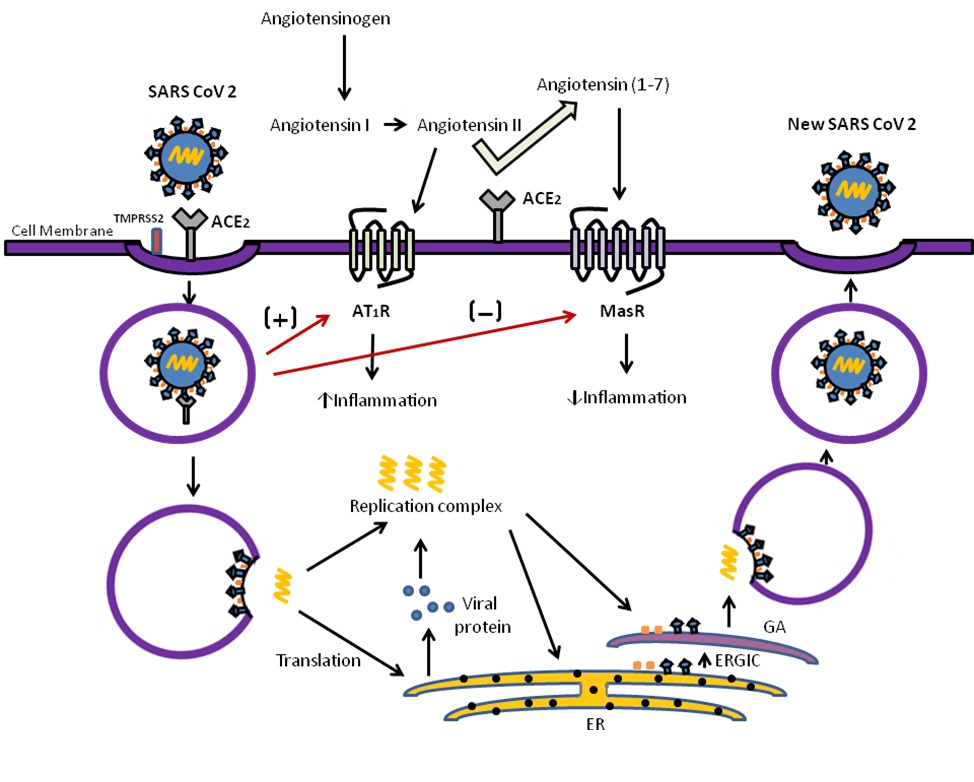
ACE2 acts as a receptor for the corona virus to enter into the cell (2). ACE-2 is a type I transmembrane metallocarboxypeptidase with homology to ACE, which plays an important role in the classical pathway of Renin-Angiotensin system (RAS). Angiotensinogen is converted in to angiotensin I and angiotensin II by the enzymes renin and angiotensin converting enzyme (ACE) respectively. Angiotensin II is involved in increasing the blood pressure by vasoconstriction, water and sodium reabsorption in kidneys, fibrosis and promote inflammation through oxidative stress by acting on AT1R (Angiotensin II receptor type 1) (12). ACE2 is the enzyme that negatively regulates the RAS by converting angiotensin I to angiotensin (1-9) and degrading angiotensin II to angiotensin-(1–7), which is a ligand for Mas receptor (MasR) (13). Angiotensin-(1–7) functions to lower the blood pressure by vasodilation, sodium and water excretion and reduce the inflammation by producing NO (Nitric oxide) (14) (Fig: 1). The ACE2 is expressed in lungs, heart, kidneys, intestine, vascular endothelial cells including alveolar epithelial type II cells (AECII), but is enriched in nasal secretary and ciliated cells (15). The AECII cells are involved in production of lung surfactant and acts as progenitor cells to alveolar epithelial type I cells (AECI) which are responsible for gas exchange.
Lungs acts as a primary target organ for SARS CoV 2 infection due to its vast surface area and harboring ability of ACE2 positive AECII cells as evidenced by expression of high levels of viral multiplication related genes (16). SARS-CoV mediated down-regulation of ACE2 receptors can cause severe inflammatory response through enhanced activation of AT1 receptors in lungs (17). In the murine apoE (Apolipoprotein E) - deficient model, ACE2 reduces the formation of aortic plaques in vivo and also restricts macrophage expression of proinflammatory cytokines in vitro, such as TNF-α and IL-6 in response to LPS (lipopolysaccharide) challenge. Thus ACE2 can directly blunt the proinflammatory response in myeloid cells (18)..
HOST CELL AND SARS-COV-2 INTERACTION
SARS-CoV-2 recognizes human ACE2 more efficiently than SARS-CoV, indicating the high rate of transmission of infection among persons (19). Cellular entry of coronavirus into the cell depends on the binding of RBM (receptor binding motif) region of S protein to the ACE2 receptor and the subsequent priming by cellular proteases like TMPRSS2 (transmembrane protease serine 2) or cathepsin B/L in lungs (9). Furin protease mediated pre cleavage of S proteins at the S1/S2 site is essential for the priming action of TMPRSS2 in lung cells (10). The binding affinity between the host receptor and the S protein determines the efficiency of viral entry, replication and finally the severity of infection. The number of ACE2 receptors are acting as a limiting factor in spread of infection, seeing that the expression level of ACE2 in different tissues including the upper respiratory tract, is less than TMPRSS2 (20). After binding, SARS-CoV-2 down regulates the ACE2 receptor function by internalizing along with it, whereas it amplifies the actions of ACE receptor, leading to severe pulmonary implications (21). Higher level of ACE2 expression on AECII is associated with the diffuse alveolar damage in SARS-CoV-2 infection leading to pulmonary edema, lung failure (22). Corona viruses have a complex genome expression strategy. After entry into the cell, RNA genome is translated into viral replicase transcriptase complex, structural and non structural polyproteins. These polyproteins are predominantly processed into functional polypeptides or protein by the virus encoded main proteases (23). Some of this proteins forms the part of the RNA-dependent RNA polymerase (RdRp) or nsp12 (non structural protein) which catalyzes the synthesis of viral RNA with the involvement of nsp7 and nsp8 as co-factors (24), while other proteins suppress the immune response to advance the viral replication and propagation. Viral protein and RNA components assemble in the Golgi to form new virions, which are transported via vesicles and released out of the cell (25).
IMMUNE RESPONSE AND CYTOKINE STORM
CONCLUSION
The emergence and rapid spread of SARS-CoV-2 arises to be a major health threat to all the countries and population around the world. The structural and functional characteristics of SARS-CoV2 explain the high rate of transmission of infection among people. Down-regulation of ACE2 receptors induced by the cell entry of SARS-CoV-2 causes severe implications of inflammatory reactions and is detrimental in pre-existing ACE2 deficiency especially in conditions of older age, hypertension, diabetes and cardiovascular disease. The interaction between the host immune system and the virus plays an essential role in onset and progress of SARS-COV-2. Further understanding of the interaction of SARS-CoV-2 and dysregulated host immune response is very essential to fight this global pandemic disease in future.
References
- De Wit, E., van Doremalen, N., Falzarano, D. and Munster, V.J., 2016. SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14(8), p.523.
- Walls, A.C., Park, Y.J., Tortorici, M.A., Wall, A., McGuire, A.T. and Veesler, D., 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell.
- Patel, V. B., Zhong, J. C., Grant, M. B., & Oudit, G. Y. (2016). Role of the ACE2/angiotensin 1–7 axis of the renin–angiotensin system in heart failure. Circulation research, 118(8), 1313-1326.
- Sodhi, C.P., Wohlford-Lenane, C., Yamaguchi, Y., Prindle, T., Fulton, W.B., Wang, S., McCray Jr, P.B., Chappell, M., Hackam, D.J. and Jia, H., 2018. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. American Journal of Physiology-Lung Cellular and Molecular Physiology, 314(1), pp.L17-L31.
- Prompetchara, E., Ketloy, C. and Palaga, T., 2020. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol, 38(1), pp.1-9.
- Zhou, Y., Fu, B., Zheng, X., Wang, D. and Zhao, C., 2020. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. National Science Review.
- Su, S., Wong, G., Shi, W., Liu, J., Lai, A.C., Zhou, J., Liu, W., Bi, Y. and Gao, G.F., 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in microbiology, 24(6), pp.490-502.
- Li, G., Fan, Y., Lai, Y., Han, T., Li, Z., Zhou, P., Pan, P., Wang, W., Hu, D., Liu, X. and Zhang, Q., 2020. Coronavirus infections and immune responses. Journal of medical virology, 92(4), pp.424-432.
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.H., Nitsche, A. and Müller, M.A., 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell.
- Hoffmann, M., Kleine-Weber, H. and Pöhlmann, S., 2020. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular Cell.
- Li, X., Geng, M., Peng, Y., Meng, L. and Lu, S., 2020. Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis.
- Sparks, M.A., Crowley, S.D., Gurley, S.B., Mirotsou, M. and Coffman, T.M., 2011. Classical renin?angiotensin system in kidney physiology. Comprehensive Physiology, 4(3), pp.1201-1228.
- Patel, V.B., Basu, R. and Oudit, G.Y., 2016. ACE2/Ang 1-7 axis: a critical regulator of epicardial adipose tissue inflammation and cardiac dysfunction in obesity. Adipocyte, 5(3), pp.306-311.
- South, A.M., Shaltout, H.A., Washburn, L.K., Hendricks, A.S., Diz, D.I. and Chappell, M.C., 2019. Fetal programming and the angiotensin-(1-7) axis: a review of the experimental and clinical data. Clinical Science, 133(1), pp.55-74.
- Sungnak, W., Huang, N., Bécavin, C., Berg, M., Queen, R., Litvinukova, M., Talavera-López, C., Maatz, H., Reichart, D., Sampaziotis, F. and Worlock, K.B., 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine, pp.1-7.
- Zhao, Y., Zhao, Z., Wang, Y., Zhou, Y., Ma, Y. and Zuo, W., Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan COVID-19. bioRxiv. 2020 Jan. Preprint]. https://doi. org/10.1101/2020.01, 26.
- Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., Huan, Y., Yang, P., Zhang, Y., Deng, W. and Bao, L., 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nature medicine, 11(8), pp.875-879.
- Thomas, M.C., Pickering, R.J., Tsorotes, D., Koitka, A., Sheehy, K., Bernardi, S., Toffoli, B., Nguyen-Huu, T.P., Head, G.A., Fu, Y. and Chin-Dusting, J., 2010. Genetic Ace2 deficiency accentuates vascular inflammation and atherosclerosis in the ApoE knockout mouse. Circulation research, 107(7), pp.888-897.
- Wan, Y., Shang, J., Graham, R., Baric, R.S. and Li, F., 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7).
- Tse, G.M., To, K.F., Chan, P.K., Lo, A.W.I., Ng, K.C., Wu, A., Lee, N., Wong, H.C., Mak, S.M., Chan, K.F. and Hui, D.S.C., 2004. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS). Journal of clinical pathology, 57(3), pp.260-265.
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X. and Cheng, Z., 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), pp.497-506.
- Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., Wang, B., Xiang, H., Cheng, Z., Xiong, Y. and Zhao, Y., 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama, 323(11), pp.1061-1069.
- Zhavoronkov, A., Aladinskiy, V., Zhebrak, A., Zagribelnyy, B., Terentiev, V., Bezrukov, D.S., Polykovskiy, D., Shayakhmetov, R., Filimonov, A., Orekhov, P. and Yan, Y., 2020. Potential COVID-2019 3c-like protease inhibitors designed using generative deep learning approaches. Insilico Medicine Hong Kong Ltd A, 307, p.E1.
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L. and Ge, J., 2020. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science.
- Shereen, M.A., Khan, S., Kazmi, A., Bashir, N. and Siddique, R., 2020. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research.
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X. and Cheng, Z., 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), pp.497-506.
- Xu, Z., Shi, L., Wang, Y., Zhang, J., Huang, L., Zhang, C., Liu, S., Zhao, P., Liu, H., Zhu, L. and Tai, Y., 2020. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet respiratory medicine, 8(4), pp.420-422.
- Zhou, P., Yang, X.L., Wang, X.G., Hu, B., Zhang, L., Zhang, W., Si, H.R., Zhu, Y., Li, B., Huang, C.L. and Chen, H.D., 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. nature, 579(7798), pp.270-273.
- Xie, B., Laxman, B., Hashemifar, S., Stern, R., Gilliam, T.C., Maltsev, N. and White, S.R., 2018. Chemokine expression in the early response to injury in human airway epithelial cells. PloS one, 13(3).
- Vaninov N. 2020. In the eye of the COVID-19 cytokine storm. Nat Rev Immunol,1:1.