Genome editing is a way of making deliberate alterations in the DNA of a cell or organism. It is basically a type of genetic engineering technique in which the DNA is precisely and efficiently inserted, deleted or modified in the genome. Genome editing has made it possible for the researchers to modify the genome of various organisms including plant, animals, and bacteria. It can be used for research purpose, to edit the genome of an organism and to understand their biology; treating human diseases and in agriculture to genetically modify crops to improve their yield and make them resistant to various abiotic and biotic factors. It is, therefore, absolutely appropriate to say that “Genome editing” has the potential to make massive changes in our lives.
How does genome editing works?
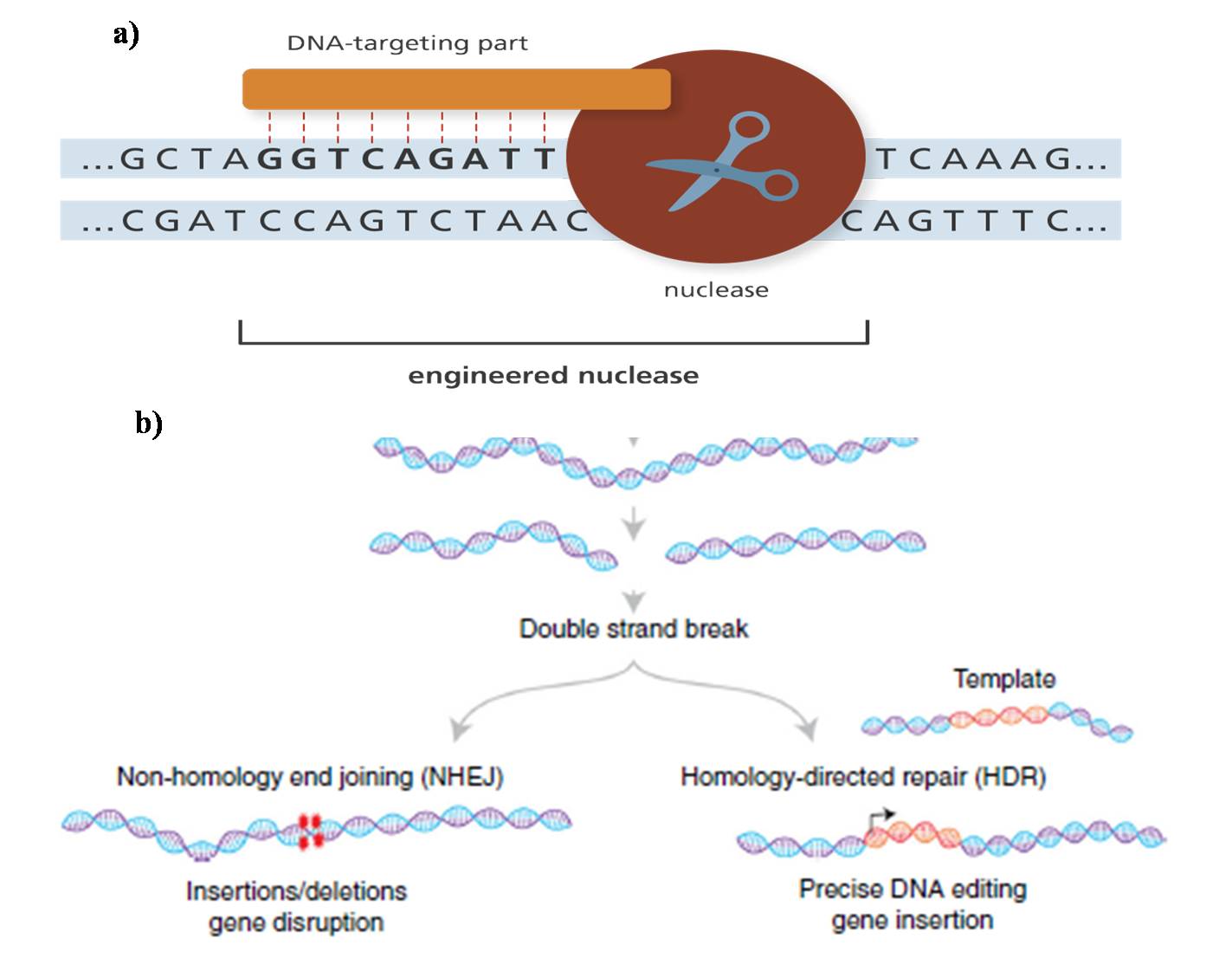
The key to genome editing is creating cuts or double-stranded breaks (DSBs) at a specific location in the genome by using engineered enzymes, the nucleases (Figure 1a). These breaks are naturally repaired by the cell through non-homologous end joining (NHEJ) or homologous recombination (HR) or Homology-directed repair finally resulting in targeted “edit” or “mutation” in the DNA sequence (Figure 1b).
The first attempt in genome editing changes was performed in yeast (1970s) and mice (1980s). This genome editing was basically dependent on the process of homologous recombination, which was highly precise but inefficient. However, the recent genome editing approach has simplified this issue, making the genetic manipulations possible in all the organisms.
Genome editing systems/approaches
The genome editing approaches marks the use of four powerful classes of engineered nucleases: (i) Meganucleases, (ii) Zinc finger nucleases (ZFNs), (iii) Transcription activator-like effector-based nucleases (TALENs), (iv) The clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system.
Meganucleases
Meganucleases began the era of genome editing and were discovered in 1985. These are endonucleases which have the potential to recognize and cut DNA sequences of 14 to 40 bp (Figure 2a). The most accepted Meganucleases are the proteins in the LAGLIDADG family. They possess a very unique feature of having a long recognition sequence (>14 bp). In fact, chances of finding accurate meganuclease for a specific DNA sequence are very low. To resolve this problem, high throughput sequencing methods have been employed to produce meganuclease variants that recognize the unique sequence. Additionally, different meganucleases were fused together to produced hybrid enzymes which are capable of recognizing new sequences.
Zinc finger nucleases (ZFNs)
Zinc finger nucleases were the first engineered nucleases which improved the effectiveness of genome editing in several ways and were discovered in 1991. The ZFNs are referred to as “genomic scissors” which makes a cut in the DNA at a specific position resulting in double-stranded breaks in the DNA molecule. ZFNs contains two main components: a) DNA binding protein domain that is designed to bind to three DNA base sequences in a sequence-specific manner, b) DNA cleaving domain which comprises of FokI nuclease which cuts the DNA. The first engineered ZFNs were prepared by binding/fusing a customized C2H2 zinc-finger protein domain to the cleavage domain of the FokI restriction endonuclease.
The DNA binding domain of ZFNs is made up of two chains of zinc-finger proteins which recognizes a 6 bp sequence each. ZFNs works by involving two DNA binding proteins with a different combination of zinc-fingers which recognizes two DNA sequences which are few nucleotides away. Binding of the two zinc finger proteins to their respective DNA sequences brings the two FokI nucleases together, which further dimerize to perform the nuclease activity and results in a double-stranded break in the DNA molecule (Figure 2b).
Transcription activator-like effector-based nucleases (TALENs)
In 2011, the second generation of engineered nucleases came into existence in the form of transcription activator-like effector-based nucleases (TALENs). TALENs comprises of two parts: a) DNA binding domain-containing transcription activator-like effector (TALE) domains, there are different TALE domains each recognizes a single nucleotide rather than a trinucleotide; b) Cleaving domain which is basically the nuclease part made up of FokI nuclease. The TALE effectors usually comprise of a repeated domain, each repeated domain contains a sequence of 34 amino acids which recognize a single DNA base within the specific target site. The nuclease FokI part of the TALENS breaks the DNA at the specific target site which is further repaired through non-homologous end-joining. Two FokI molecules are required to create double-stranded breaks in the DNA molecule, therefore two TALENS are required one for each DNA strand. (Figure 2c).
TALENS can be easily designed and prepared in a short span of time compared to ZFNs. They can be easily targeted across the genome. They have less off-target cytotoxic effects on the host cells. On the contrary, they are large in size compared to the ZFNs and hence difficult to be delivered in vivo.
The clustered regularly interspaced short palindromic repeats (CRISPR-Cas9) system
CRISPR-Cas9 has become a revolutionary genome editing technology in biological research because it is robust, enables precise genetic modifications and displays high editing efficiency than the other editing tools. This method promises to dramatically improve our ability to edit the genome of any species including humans. In 2012, Jennifer Doudna, Emmanuelle Charpentier, and their teams reported for the first the mechanism of CRISPR technology. CRISPR-Cas9 comprises of two components: a) CRISPR stands for clustered regularly interspaced short palindromic repeats which are the DNA-targeting part of the system; b) Cas9 stands for CRISPR-associated protein which is the nuclease part of the system (Figure 2d).
CRISPR-Cas9 was adapted from a naturally occurring system in bacteria used to protect themselves from invading viruses. The bacteria detect the presence of virus DNA and immediately produce two short RNA sequences known as guide RNAs (gRNAs), one of which contains sequence matching that of the viral DNA. These two RNAs form a complex with Cas9 protein, the nuclease part that cuts the DNA. When the guide RNA finds its target within the viral genome, Cas9 cuts the target DNA disabling the virus. Over past few years, researchers working on this system have realized that it can be engineered to cut not just the viral DNA but any DNA sequence at a precise location by changing/modifying the guide RNA sequence so as to match the target DNA. This can be done no just in the laboratory but inside the nuclease of the living cell. The short synthetic RNA segments are created which contains a short “guide” sequence which binds with the Cas9 enzyme and a user defined ~ 20 nucleotide spacer that defines the genomic target to be modified. The guide RNA binds to the specific target DNA sequence in the genome. Once the guide RNA recognizes the target sequence, Cas9 nuclease cuts the DNA at the precise location. Once the DNA is cut, the cell uses its own repair machinery to add or delete the DNA fragment resulting in errors to generate a gene knockout, or additional genetic modifications are introduced by replacing the existing DNA segment with a customized DNA sequence. CRISPR-Cas9 technology is no longer a genome editing tool, rather it is now applied in targeted gene regulation, epigenetic modifications, chromatin engineering. Unlike previous editing methods, CRISPR can be used to target many genes at once, a significant advantage to study complex human diseases that are caused by many genes acting together.

Figure 2. Genome editing approaches a) Meganucleases; b) Zinc finger nucleases (ZFNs); c) Transcription activator-like effector-based nucleases (TALENs); d) The clustered regularly interspaced short palindromic repeats (CRISPR-Cas9) system. (Source: Gaj et al., 2016; Ding et al., 2016).
Applications of Genome editing
Genome editing techniques can be used for diverse range of applications across science, biotechnology and medicine. In science, the editing tools have been used to study gene function. In biotechnology, these technologies have been applied to agriculture, to genetically modify the plant genome to confer resistance to abiotic and biotic stresses as well as to improve their yields. In medicine, the genome editing tools are used to treat human diseases. These techniques enable manipulation of the human genome sequence and have tremendous potential to treat inherited diseases. They can improve cell therapies that are designed to overcome cancer and regenerate damaged tissues.
References
1.Gaj T., Sirk SJ., Shui SL., Liu J. (2016). Genome-Editing Technologies: Principles and Applications. Cold Spring Harbor Perspectives in Biology, 8:a023754.
2.Germini D., Tsfasman T., Zakharova VV., Sjakste N., Lipinski M., Vassetzky Y. (2017). A comparison of techniques to evaluate the effectiveness of genome editing. Trends in Biotechnology, 36: 147-159.
3. Adli M. (2018). The CRISPR tool kit for genome editing and beyond. Nature Communications, 9:1911.
4. Genome editing in brief: what, why and how? (http://nuffieldbioethics.org/report/genome-editing-ethical-review/genome-editing)
5. What is genome editing? (https://www.yourgenome.org/facts/what-is-genome-editing)
6. CRISPR Cas9. (https://www.horizondiscovery.com/gene-editing/crispr-cas9)