The pandemic- COVID-19, has completed its one year since its inception from the sea food market of Wuhan, China. Till to date this has taken 2.75 million human lives all over the world. Scientists are working round the clock to come with an effective vaccine. Till to date 13 number of vaccines have been approved/ authorized. So far the scientific studies suggest the virus, SARS-CoV-2(the cause of COVID-19), likely had ancestors that originated in bats, followed by transmission to an intermediate host, and that both viruses may have an extended host range that includes primates and other mammals.
Many mammalian species host coronaviruses and these infections are frequently associated with severe clinical diseases, such as respiratory and enteric disease in pigs and cattle. Additionally, Molecular phylogenetics revealed that at least one human coronavirus (HCov-OC43) may have originated in cattle or swine and that this virus was associated with a human pandemic that emerged in the late 19th century.
Recent data suggest that coronaviruses can be transmitted from bats to other wildlife species and humans, and from humans to tigers and pigs. Hence, understanding the host range of SARS-CoV-2 and related coronaviruses is essential for improving our ability to predict and control future pandemics. It is also crucial for protecting populations of wildlife species in native habitats and under human care, particularly nonhuman primates, which may be susceptible to COVID-19. An excellent study published in the journal “Proceedings of the National Academy of Sciences of the United States of America” in August, 2020 revealed some interesting findings that support the above paragraph.
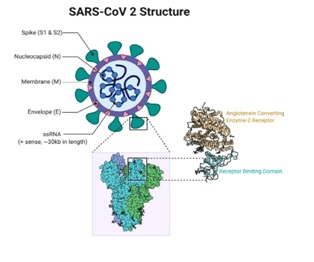
How this SARS-CoV-2 starts its journey into cells? Coronavirus entry into host cells is an important determinant of viral infectivity and pathogenesis. To enter host cells, coronaviruses first bind to a cell surface receptor for viral attachment, subsequently enter endosomes, and eventually fuse viral and lysosomal membranes. A virus surface-anchored spike protein mediates coronavirus entry. The angiotensin I converting enzyme 2 (ACE2) serves as a functional receptor for the spike protein (S) of SARS-CoV and SARS-CoV-2. Under normal physiological conditions, ACE2 is a dipeptidyl carboxypeptidase that catalyzes the conversion of angiotensin I into angiotensin 1-9, a peptide of unknown function. ACE2 also converts angiotensin II, a vasoconstrictor, into angiotensin 1-7, a vasodilator that affects the cardiovascular system and regulate other components of the renin–angiotensin system.
What is the essence of this study?
Coronaviruses may adapt to new hosts in part through mutations in S that enhance binding affinity for ACE2. The best-studied example is the evolution of SARS-CoV-like coronaviruses in the masked palm civet, which is believed to be the intermediate host for transmission of a SARS-CoV-like virus from bats to humans. The masked palm civet SARS-CoV S acquired two mutations that increased its affinity for human ACE2. Hence, to provide insights for the future zoonotic transmissions and to predict which vertebrate species could be the potential harbour of this pathogen, scientists took the help of genomics. They used a combination of comparative genomic approaches and protein structural analysis to assess the potential of ACE2 homologs from 410 vertebrate species (including representatives from all vertebrate classes: fishes, amphibians, birds, reptiles, and mammals) to serve as a receptor for SARS-CoV-2 and to understand the evolution of ACE2/SARS-CoV-2 S-binding sites. Interestingly their findings reinforced earlier findings on the natural host range of SARS-CoV-2 and predict a broader group of species that may serve as a reservoir or intermediate host(s) for this virus. Importantly, many threatened and endangered species were found to be at potential risk for SARS-CoV-2 infection based on their ACE2 binding score, suggesting that as the pandemic spreads humans could inadvertently introduce a potentially devastating new threat to these already vulnerable populations, especially the great apes and other primates. The following tables are categorized on the basis of ACE2/SARS-CoV-2 S-binding sites. This shows the risk level of species susceptible for SARS-CoV-2.
|
HIGH |
Common Name |
Scientific Name |
IUCN Status |
|
Human |
Homo sapiens |
|
|
|
Western lowland gorilla |
Gorilla gorilla gorilla |
CR |
|
|
Northern white-cheeked gibbon |
Nomascus leucogenys |
CR |
|
|
Sumatran orangutan |
Pongo abelii |
CR |
|
|
Proboscis monkey |
Nasalis larvatus |
EN |
|
|
Bonobo |
Pan paniscus |
EN |
|
|
Chimpanzee |
Pan troglodytes |
EN |
|
|
Red-shanked douc |
Pygathrix nemaeus |
EN |
|
|
Golden snub-nosed monkey |
Rhinopithecus roxellana |
EN |
|
|
Sooty mangabey |
Cercocebus atys |
EN |
|
|
Southern pig-tailed monkey |
Macaca nemestrina |
VU |
|
|
Rhesus monkey |
Macaca mulatta |
LC |
|
|
Drill |
Mandrillus leucophaeus |
EN |
|
MEDIUM |
Common Name |
Scientific Name |
IUCN Status |
|
Aye-aye |
Daubentonia madagascariensis |
EN |
|
|
Syrian hamster |
Mesocricetus auratus |
VU |
|
|
Sperm whale |
Physeter catodon |
VU |
|
|
Barbary sheep |
Ammotragus lervia |
VU |
|
|
Bison |
Bison bison bison |
NT |
|
|
Hirola |
Beatragus hunteri |
CR |
|
|
Wild yak |
Bos mutus |
Domesticated |
|
|
Wild goat |
Capra aegagrus |
NT |
|
|
Masai giraffe |
Giraffa tippelskirchi |
EN |
|
|
Nilgiri tahr |
Hemitragus hylocrius |
EN |
|
|
Sunda clouded leopard |
Neofelis diardi |
VU |
|
|
Jaguar |
Panthera onca |
NT |
|
|
Leopard |
Panthera pardus |
VU |
|
|
Siberian tiger |
Panthera tigris altaica |
EN |
|
|
Cheetah |
Acinonyx jubatus |
VU |
|
|
Hippopotamus |
Hippopotamus amphibius |
VU |
|
|
Dama gazelle |
Nanger dama |
CR |
|
|
European rabbit |
Oryctolagus cuniculus |
EN |
|
LOW |
Common Name |
Scientific Name |
IUCN Status |
|
Southern white rhinoceros |
Ceratotherium simum simum |
NT |
|
|
Black rhinoceros |
Diceros bicornis |
CR |
|
|
Giant panda |
Ailuropoda melanoleuca |
VU |
|
|
Wild Bactrian camel |
Camelus ferus |
CR |
|
|
Polar bear |
Ursus maritimus |
VU |
|
|
Bush dog |
Speothos venaticus |
NT |
|
|
Brownhead whale |
Balaena mysticetus |
LC |
|
|
African elephant |
Loxodonta africana |
VU |
|
|
Straw coloured fruit bat |
Eidolon helvum |
NT |
|
|
Large flying fox |
Pteropus vampyrus |
NT |
|
|
Patagonian mara |
Dolichotis patagonum |
NT |
|
|
Gray mouse lemur |
Microcebus murinus |
LC |
Analysis of this study
Out of 401 vertebrate species taken into consideration in this study, 103 species that scored very high, high, and medium for ACE2/SARS-CoV-2 S binding, 41 (40%) are classified in one of three “threatened” categories (vulnerable, endangered, and critically endangered) on the International Union of Conservation of Nature (IUCN) Red List of Threatened Species, five are classified as near threatened, and two species are classified as extinct in the wild. For example in Cetacea, 12 of 14 species score as high, and of those two are threatened. Toothed whales have potential for viral outbreaks and have lost function of a gene that is key to the antiviral response in other mammalian lineages. If they are susceptible to SARS-CoV-2, human-to-animal transmission could pose a risk through sewage outfall and contaminated refuse from cities, commercial vessels, and cruise liners.
Conclusion
Although this in silico studies suggest potential susceptibility of diverse species, verification of infection potential is required, using cell cultures, stem cells, organoids, and other methods that do not require direct animal infection studies. Zoos and other facilities that maintain living animal collections are in a position to provide such samples for generating crucial research resources by banking tissues and cryo-banking viable cell cultures in support of these efforts.
References
1) Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates, Damaset al., Proceedings of the National Academy of Sciences of the United States of America, August, 2020.https://doi.org/10.1073/pnas.2010146117. 2)https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker