Nanopore-based sequencers is a fourth-generation DNA sequencing technology. Nanopore is a hole with size range in nano which is created by eitherproteins present in lipid bilayer membrane (biological nanopores) or poreof solid materials origin (solid-state nanopores)or hybrid pore including both.Idea of nanopore sequencing was proposed by Deamer and Branton and independently by Church in the 1990s.it was proposed that nanopore can be used to sequence DNA/RNA molecules by applying external voltage.Nanopore-based sequencing technologies have originated from the Coulter counter and ion channels.
Why Nanopore? Nanopore technology offers several aids over other sequencing technology, such as:
- High Sensitivity
- Ability to sequence single molecules
- Long read lengths
- Real-time analysis
- Cost-effectiveness
- Low sample requirement
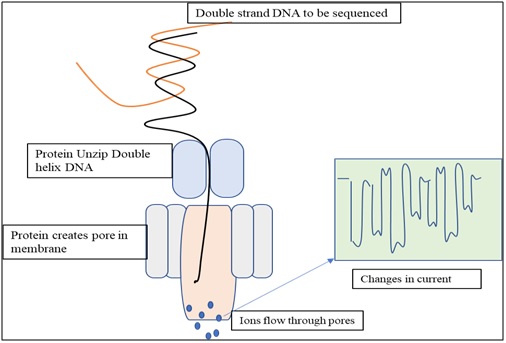
Principal of nanopore sequencing technology: Principal of nanopore sequencing relies on changes in ionic currents during translocation of DNA via a tiny channel with pore size in nm range. DNA to be sequenced is passed through a microscopic pore in lipid bilayer membrane. One protein opens the dsDNA helix in to two strands. Second protein creates a pore in to the membrane. Flow of ions through the pore creates a current. Bases are identified by the way they affecting the flow of ions through the pores. Each base block/ affect the ion flow to a different degree and the degree of electric current disruption by the corresponding base are measured.
Types of Nanopore: There are three types of nanopores are used
- Biological nanopores (Protein embedded in lipid bilayer
- Synthetic or Solid state nanopores (solid substrates are formulated in to defined pore size)
- Hybrid nanopore (Hybrid of biological and synthesis molecules)
Biological Nanopore: Biological nanopores are also called as transmembrane protein channels, which arearegenerally inserted into planar lipid bilayers structure, liposomes, andother lipid-based polymer films. First experiments on translocation of biopolymer (DNA) across the lipid bilayer was conducted by Deamer, Branton, and Kasiannowicz in 1993 using α-hemolysin (α-HL). Biological nanopore allows to pass molecules (ssDNA or RNA) through the channel diameter (1.5 nm). Example of biological nanopores are- a-Hemolysin, MspA and Phi 29.
Advantage: Size and structure of biological nanopores are well-defined and highly-reproducible. Biological nanopores can be easily manipulated. Using advance molecular biology techniques mutation in nucleaotide sequence can be done to change the amino acid residue at a specific position.
Disadvantage: With ease in manipulability, biological nanopores have disadvantages too. Because of biological origin they are fragile in nature. Limited pore size has restricted its application in small molecule analysis, ssDNA, etc. Biological activity and stability of nanopore depends on various environmental conditions like, temperature, salt concentration, pH and others.
- α- Hemolysin:The first and most commonly biological nanopore used for DNA sequencing is α-Hemolysin. It is an exotoxinsecreted by the bacterium Staphylococcus aureus which is a human pathogen.It is aheptameric (mushroom-shaped) transmembrane channel structure with molecular weight 232.4-kDa, containing a 3.6-nm diameter cap and a 2.6-nm diameter transmembrane barrel structure. The external dimensions of the pores are 10 nm · 10 nm.α -Hemolysin has ability to pass and detect various molecules such as metal ions, DNA, RNA, Protein and small organic molecules possessing diameter of ~ 1 nm. They are thermally stable and functions even at higher temperature ie, 1000C and work on wide ph range (pH 2–12)
- MspA (Mycobacterium smegmatis porin A): It is most powerful and commonly used biological nanopore after α- Hemolysin. The narrow point is 1nm in diameter comparatively smaller that α- Hemolysin allows spatial resolution of SSDNA. Alike α- Hemolysin, MspAfuctions on broad range of Ph 0-14. It has been reported that site directed mutagenesis can also be performed in this nanopore. It was observed that MspA nanopore shows better base resolution than α-HL nanopore by creating larger signal difference between the bases.
- Bacteriophage phi29. α- Hemolysin and MspA allows to pass the molecules below 1.5 nm diameter, hence these nanopores are not useful for dsDNA Molecule. Gua et al., 2009 first proposed the insertion of phi29 connector protein in lipid bilayer. The channel consists of twelve copies of gp10 protein, which encircle to form a dodecamer channel. The length of this connector is ~7 nm, while the cross-sectional area of the channel is 10 nm2 (larger diameter 3.6 nm allows translocation of dsDNA molecules) at the narrow end and 28 nm2 (6 nm in diameter). Bacteriophage phi29 shows higher conductance than previous two biological nanopore
Synthetic nanopores:
Biological nanopores are good for molecule with smaller diameter because of their constant pore size and susceptible to extreme environmental conditions as explained above which leads to affect the stability. To overcome the problems associated with biological nanopores, synthetic nanopores have been designed using various solid support material via different fabrication process. Therefore, we can design or adjust the pore size and increased the thermal and mechanical stability of synthetic nanopores. Chemical, Thermal and mechanical stability of pore depends on the fabricated material and method of fabrications. Various method of fabricating nanopores are as follows-
- Ion milling track-etch method
- Focused ion beam method
- Electron beam based decomposition sputtering method
- Focus ion bean method
- Laser ablation method
- Electron-beam lithography method
- Helium ion microscopy method
- And the latest dielectric breakdown method
Solid-state nanopores have been introduced in various fields, such as DNA sequencing, detection of protein and molecules, molecule translocation process, and disease diagnosis. Most common material used in synthetic nanopores are silicon nitride (Si3N4) silicon dioxide (SiO2), aluminum oxide (Al2O3) boron nitride (BN), graphene, polymer membranes etc.
Disadvantage: Synthetic nanopore do not differentiate the target and artifacts of same size. hence the lack of chemical differentiation is one of the major drawbacks of solid state nanopores.
Hybrid Nanopore: Hybrid nanopore, as the name indicate are the hybrids of biological and synthetic nanoporehas attracted growing interests due to its excellent conductivity, atomic thickness, and robust mechanical stability. Limitation of synthetic nanopore to differentiate between the molecule of same size can be overcome by hybrid materials. Specificity towards chemicals can be amended by surface functionalization or by conjugating specific recognition sequences to the site of nanopore. Synthetic nanopores can be subjected for coating with lipid bilayer in a controlled manner to regulate the process of protein translocation
Biological vs Synthetic nanopores
|
Properties |
Biological nanopore |
Synthesis nanopore |
|
Pore stability |
Not stable: susceptible to extreme Ph, salt concentration, and temperature |
Highly stable: Stable chemical thermally and mechanically |
|
Channel size |
Constant and usually smaller: MspA provide better spatial resolution as compared to α-hemolysin. |
Smaller to higher |
|
Fragility |
Very high, due to traditionally supported lipid membranes |
Stable due to solid support andfabricated materials |
|
Surface functionalization |
Relatively easy |
Not so easy |
|
Modification |
site directed mutagenesis can be made easily to alter the size and charge properties of the channel |
Difficult because synthetic pore lack chemical and location selectivity
|
|
Production |
Easy |
Labor intensive |
|
size adjustability |
Not possible |
Possible |
|
Cost |
Biological pores can be harvested in cells in large quantities at low cost. |
Costly as compared to biological nanopores |
References
Deamer, D. W., &Branton, D. (2002). Characterization of nucleic acids by nanopore analysis. Accounts of chemical research, 35(10), 817-825.
Wang, Y., Yang, Q., & Wang, Z. (2015). The evolution of nanopore sequencing. Frontiers in genetics, 5, 449.
Agah, S., Zheng, M., Pasquali, M., &Kolomeisky, A. B. (2016). DNA sequencing by nanopores: Advances and challenges. Journal of Physics D: Applied Physics, 49(41), 413001
Haque, F., Li, J., Wu, H. C., Liang, X. J., & Guo, P. (2013). Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano today, 8(1), 56-74.
Feng, Y., Zhang, Y., Ying, C., Wang, D., & Du, C. (2015). Nanopore-based fourth-generation DNA sequencing technology. Genomics, proteomics & bioinformatics, 13(1), 4-16.