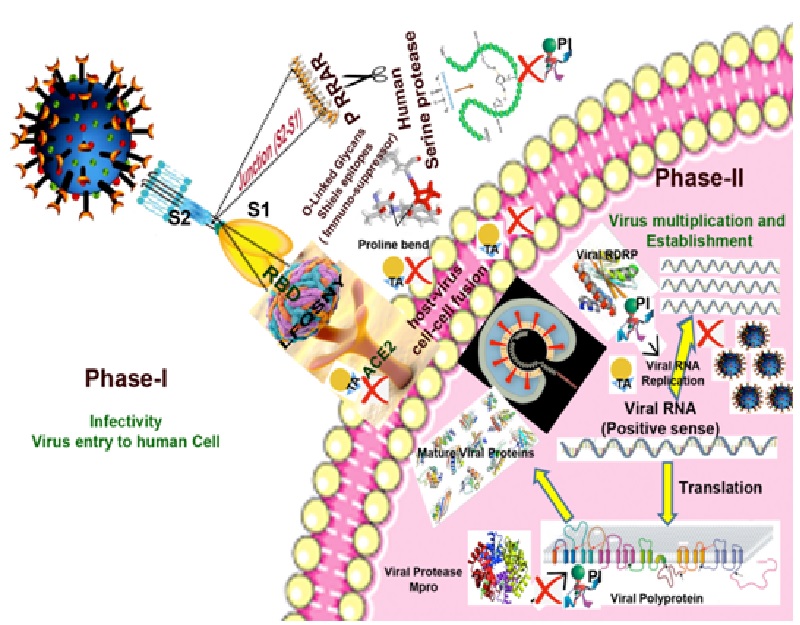
Coronavirus disease-2019 (COVID-19) pandemic has inflicted more than 2.85 millions infections with a mortality of 1,96,295 as on 27th April 2020. SARS-CoV-2 has been identified as the etiological agent of viral pneumonia pandemic which had its origin in Wuhan, China. The key viral proteins are: spike (S) glycoproteins having role in infection process, main protease (Mpro) with a role in the maturation and processing of viral proteins and RNA-dependent RNA polymerase (RdRp)-which is involved in the viral replication1.
Unlike other coronaviruses, spike protein (S) of SARS-CoV-2 are efficient in interacting with human angiotensin-converting enzyme2 (hACE2) receptor. Human serine proteases like ‘Furin’ further potentiate the membrane fusion capability of the virus. In this communication we present the biochemical features of early and late infection stages of virus and analyze the available candidate therapeutics and to suggest the suitable diagnostic tools.
Biochemical features of viral proteins and enhanced pathogenicity:
The rapid spread of SARS-CoV-2 could be attributed to the unique mutations in the gene sequence of spike (S) glycoproteins. The possible modifications in S protein that broadened the host range for this virus are: (i) acquire the ability to get activated by the host proteases and (ii) its binding efficacy with the surface human receptor proteins. Spike protein sequence of SARS-CoV-2 has acquired modulations by developing: (i) a polybasic catalytic site to human serine protease ‘Furin’, which potentiates the membrane fusion capabilities of the virus. (ii) high affinity of S protein-Receptor Binding Domain (RBD) towards human cell receptor ACE2 owing to six amino acids viz., L455, F486, Q493, S494, N501, Y505 in general and N501T mutation from SARS-CoV3.
Human serine protease-Furin
Furin, a human serine protease, is ubiquitously expressed as transmembrane protein. It cleaves a plethora of proteins at polybasic recognition motifs and releases the bioactive counterparts in the secretory pathway. In a normal cell, Furin catalyzes the activation of human receptors, hormones, zymogens, and cell surface proteins. Incidentally, most viral envelope glycoproteins also require to be proteolytically cleaved prior to entry into host cells. Quite interestingly, the S protein of SARS-CoV-2 carries a poly basic P681RRAR at the junction of its S1 and S2 subunits. The conformational bend created by the proline at the position 681 is predicted to cause the addition of O-linked glycans to amino acids which flank the cleavage site and are unique to SARS-CoV-2. O-linked glycans shield the epitopes and helps the virus in evading the host immunity.
Role of viral proteins after cell entry
SARS-CoV-2 utilizes the polyprotein strategy of genome information processing. These polyproteins are predominantly processed into functional polypeptides by the virus encoded main protease (Mpro). Further, studies on the structural backbone and active site conformation of selected Mpro disclosed that despite the subtle sequence variations among the viruses, the overall structural integrity and active site conformation are maintained4. Another crucial protein of viral origin is RNA-dependent RNA polymerase (RdRp), which is involved in the generation of viral RNA copies from the RNA templates. Coranaviruses utilize multi-subunit replication/transcription machinery and the central key component of this set up, nsp12 (RdRp) which is involved in replication of viral RNA could also be a potential drug target.
Table 1. List of therapeutic agents (TA) and viral protein inhibitors (PI) as potential drugs to treat COVID-19 disease6-10
|
Sl.No |
Point of action |
Drug and analogues |
Possible mode of action |
|
1 |
Virus entry (Phase-I) |
Arbidol (TA), Baricitinib (TA), Camostatmesylate (PI), Chloroquine, hydroxychloroquine (TA), EK1 (TA), Human recombinant soluble ACE2 (TA), Nafamostat (TA), Niclosamide (TA), Teicoplanin (TA) |
Inhibits viral entry, endocytosis, membrane fusion, and modulates endosomal pH
|
|
2 |
Protease inhibitors (Phase-II) |
Alpha-ketoamide inhibitors (PI), Chlorophenyl-pyridyl-carboxamide derivative (PI), Danoprevir (PI), Darunavir (PI), Ritonavir and Lopinavir (PI) |
Inhibitor of Mpro and other viral endopeptidase C30 |
|
3 |
RNA dependent RNA polymerase inhibitors (Phase-II) |
Alovudine and AZT (TA), Favipiravir (PI) Remdesivir (PI), Ribavirin (PI), Theaflavin (PI) |
Terminates polymerase extension in SARS-CoV and inhibit RdRp
|
Candidate drugs to treat COVID-19
The on-going clinical trials and data indicate that there is a need for a molecule or consortia of drugs that can target multiple viral proteins at different levels (Table 1). At the initial phase (Phase-I) the potential drug targets are inhibitors of serine proteases like Furin and modulation of receptor proteins like ACE2. At later stages (Phase-II and further), once the virus enters the host cell, targeting the viral non-structural proteins (nsps) like Mpro and RdRp-which are essential for the survival and replication of virus seems a viable approach (Table 1). The potency of plant botanicals and metabolites in suppressing the viral proteins also indicate the importance of diet in fighting COVID-19. Also in the disease diagnostic front, tests for SARS-CoV-2 have revealed that in the early stage of infection RT-PCR has been the method of choice whereas during the late phase total antibody, IgM and IgG –based protein detection methodologies are most suitable5.
Conclusions
We have provided a biochemical overview of COVID-19 infection process by highlighting the molecular arsenal of SARS-CoV-2 (Fig.1). At the phase-I, during the virus entry to the host cell, the balance of power is heavily shifted in favour of the pathogen. In Phase-II, viral Mpro processes the mature viral proteins required for maintenance and establishment along with the viral RdRp in performing the replication of the virus. Hence, in order to counter the SARS-CoV-2 infection, a multi-pronged strategy of utilizing various therapeutic agents to arrest the Spike-protein-mediated entry of virus in Phase-I and protein inhibitors to impede the Mpro and RDRP mediated virus maintenance and propagation during Phase-II and later stages is envisioned (Fig.1).
Disclaimer: The views expressed in the article are those of the authors and not of the organization they belong to
Acknowledgement:
Critical suggestions and sharing of related articles by Dr.P.UshaSarma, Former Scientist from Molecular Biochemistry and Diagnostics, Institute of Genomics and Integrative Biology (IGIB), New Delhi, India is hereby acknowledged.
References:
- Srinivasan, S., et al., Viruses 2020, 12, 360. doi:10.3390/v12040360
- Wan Y., et al., J. Virol. 2020, 94:e00127-20. https://doi .org/10.1128/JVI.00127-20
- Andersen K.G. et al. Nat Med 2020. https://doi.org/10.1038/s41591-020-0820-9
- Zhang L. et al. Science, 2020, 3405, 1–9.
- Zhao, J. et al. Clin. Infect. Dis. 2020, https://academic.oup.com/cid/advance article/doi/10.1093/cid/ciaa344/5812996
- Deng, L., et al., . J Infect. 2020 https://www.sciencedirect.com/science/article/pii/S0163445320301134
- Richardson, P., et al., Lancet 2020, https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30304-4/fulltext
- Lin S.,et al., 2020, https://doi.org/10.11.1/2020.01.31.92965
- Cao, B., et al. N Engl J Med. 2020, https://www.nejm.org/doi/full/10.1056/NEJMoa2001282
- Manli, W., et al., Cell Research. 2020, 0:1-3.