Artificial Intelligence is transformings every field of science. DeepMind, a 12-year-old AI-based London company, has achieved many breakthroughs in computer games, neuroscience, engineering, mathematics, and environmental science through artificial intelligence (AI). For instance, DeepMind developed an AI tool for a faster way of diagnosing and understanding diabetic retinopathy and age-related macular degeneration, which affected over 100 million people causing permanent sight loss. On July 28, 2022, the company announced its recent triumph: DeepMinds’ AlphaFold had just constructed a massive catalogue for the 3-D structure of all known curated proteins on earth.
Why is it so important?
Proteins are the fundamental molecular machines that perform almost every function in the cell. The specific sequence of 20 different amino acids determines protein 3-D structure. The folded form provides a suitable shape that places atoms of essential residues in appropriate space and time to execute the molecular function. Any undesirable perturbation to protein function such as cancer mutations, viral infections and binding with unfavourable molecules can result in human diseases. The knowledge of 3-D structure helps us to understand the function of proteins and guides us to engineer proteins for therapeutic, diagnostic and biotechnological purposes.
In 1957, the very first structure was released for a protein (myoglobin), and it was determined using X-ray crystallography by the strenuous effect of John Kendrew at Cambridge University. Since then, experiments namely X-ray crystallography, nuclear magnetic resonance and cryo-electron microscopy techniques have been applied to determine the 3-D structures of proteins. As such the experimental approaches require immense time, effort, money and energy. Also, some proteins are simply not amenable to experimental conditions for their structure determination. Efforts have been made to predict 3-D structures in computers without any experiments. For example, an algorithm called ‘Modeller’ developed by Andrej Sali and Tom L Blundell at Birkbeck College, London, predicts protein structures using available experimental structures obtained from their cousin proteins (i.e. evolutionarily related). Despite significant contributions of these approaches, the structural space didn’t cope with the exponential growth of sequence space obtained from rapid next-generation sequencing of whole genomes until recently.
AlphaFold and AlphaFoldDB
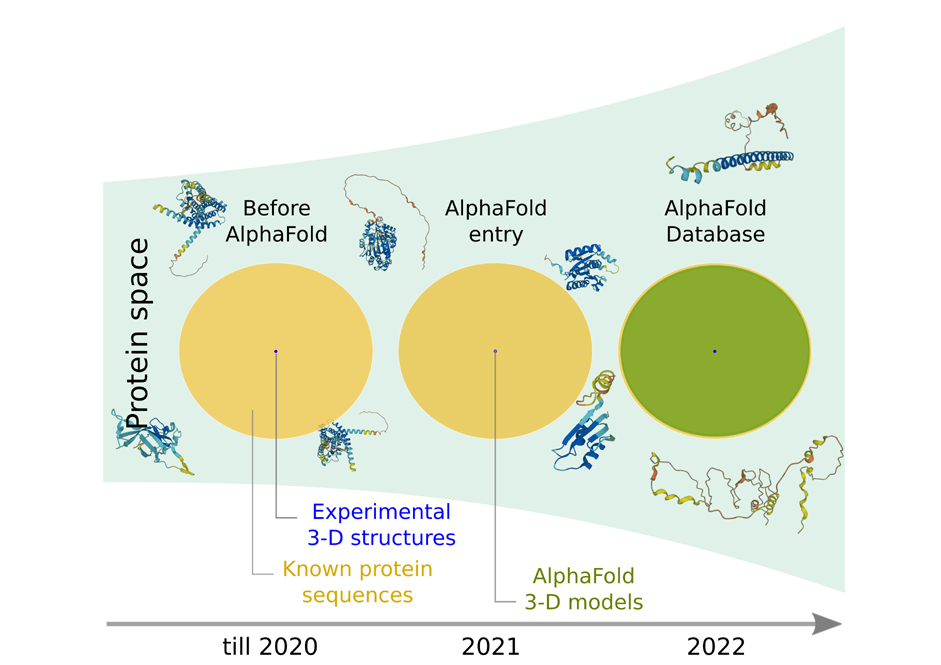
In 2018, DeepMind’s AlphaFold entered the field of structural biology (as A7D) and predicted protein structures with startling accuracy. It achieved so by iteratively learning and relearning the patterns of amino acid interactions in ~ 1 million experimentally solved structures and sequence conservation. In two years, its updated version, AlphaFold 2 showed a remarkable performance by outcompeting the previous version as well as 100 existing structure prediction tools. Within months, it made a gigantic leap by generating 3-D models for 3,50,000 proteins that covered 44% of human genomes and twice the number of experimental structures. This year in collaboration with EMBL’s European Bioinformatics Institute (EMBL-EBI), AlphaFold has generated 200 million structures covering all known proteins catalogued in the UniProt, a comprehensive protein database.
The models are available in AlphaFold Protein Structure Database, AlphaFoldDB. The database provides new tools where you can look up the 3D structure of a protein almost as easily as you can do a keyword Google search, the remark made by Demis Hassabis, founder and CEO of DeepMind at a press conference in London. The power of AlphaFold giving 3-D models within just 10 to 20 seconds per protein will foster upcoming novel discoveries in digital biology.
AlphaFold-aided scientific developments
AlphaFold had already well integrated into the field and aided structural biologists in developing a vaccine candidate for malaria, engineering proteins for recycling single-use plastics, and studying disease resistance proteins. Arne Elofsson's group at Stockholm University, Sweden demonstrated that AlphaFold improves the prediction of protein-protein interactions. AlphaFold motivates computational biologists to develop different application tools. For example, ColabFold predicts structures much faster for individual proteins as well as proteins assembled in a complex form with the help of AlphaFold. ColabFoldDB minimises the storage space and at the same time expands the resource required for AlphaFold to run by covering millions of additional protein sequences obtained from environmental metagenomic samples. Such an effect enables us to detect novel proteins from microbes in the environment and rapidly diagnose pathogens in water or clinical samples. The collaboration between Johannes Soeding at Max Planck Institute for multidisciplinary Sciences, Germany and Martin Steinegger at Seoul National University, South Korea groups resulted in a tool called FoldSeek that finds structurally similar proteins in the AlphaFoldDB with the speed ever achieved before. This is a crucial achievement that accelerates many protein bioinformatics analyses, for example, identifying drug-target proteins in pathogens that don’t perturb human proteins and evolutionary links between living and extinct species.
Protein Structural Biology in India
India has been playing a significant role in the protein structural biology field starting from the discovery of the Ramachandran map by G. N. Ramachandran to eminent crystallographic works by G. Kartha on ribonuclease and M. Vijayan on insulin and lectins. India has many well-equipped facilities for protein structure determination experiments in renowned institutes such as IISc, IITs, IISERs and TIFRs supporting Indian researchers to contribute to the growth of 3-D structure information significantly. In 2017, the country established the first cryo-EM facility in NCBS, Bangalore. We now have four cryo-EM facilities around the country including one recently opened in CCMB, Hyderabad. The Science and Engineering Research Board (SERB), an institution under the Department of Science and Technology, India has taken responsibility for providing support that will allow Indian researchers to take the lead in the cryo-EM field. This is evidenced by recent achievements in designing peptide inhibitors for SARS-CoV-2 spike protein and revealing the mechanism of actin filament using the cryo-EM technique. In addition to academic labs, separate organisations like the Institute of Bioinformatics and Applied Biotechnology, Bangalore incubates nearly 20 commercial companies and offers summer internships, diploma, master and PhD programs to foster applied bioinformatics research. The Bioinformatics, AI and Big Data programme along with the establishment of Bioinformatics and Computational Biology Centres by the Department of Biotechnology, India promotes cutting-edge research involving large-scale biological data and Artificial Intelligence.
The AlphaFoldDB covering the whole proteome of 48 organisms that are important for research and global health will be a treasure trove for scientists in India and around the world. We can foresee that scientific advances benefiting from this database and AI will pave paths not only in developing protein structural biology fields but also in promoting more specialised research programmes to create further career opportunities. It may result in funding by the government and industries to encourage AI research projects at the interface with biology, which will have a positive impact on medicine. Overall, this is undoubtedly a moment to celebrate the breakthrough made by Deep Mind’s AlphaFold and hope for many scientific and biotechnological inventions in India.
Acknowledgements
Dr. Johannes Soeding, Max Planck Institute for Multidisciplinary Sciences, Germany and the late Prof. N. Srinivasan, Indian Institute of Science, Bangalore, India.