ChIP-Seq or ChIP-Sequencing is a powerful technique to study protein-DNA interactions. ChIP-Seq is a combination of chromatin immunoprecipitation (chIP) with massively parallel next-generation DNA sequencing. It allows identification of DNA binding proteins mainly transcription factors, chromatin-modifying proteins and histones associated proteins influencing phenotype in a genome-wide manner.
Identification of how proteins interact with DNA to regulate gene expression is highly significant and informative for a better understanding of various biological processes and diseases. ChIP-Seq is primarily evolved from ChIP-chip assay which requires hybridization assay. In the past years, the rapid advancements in next-generation sequencing (NGS) techniques have allowed researchers to sequence millions of short DNA fragments in a single run. ChIP-Seq was one of the early applications of NGS and the technique was first described in 2007. In comparison to ChIP-chip (microarray) assay, ChIP-Seq provides superior quality data owing to its various attributes such as high resolution, greater genomic coverage, fewer artifacts and broad dynamic range.
How do ChIP-Seq works?
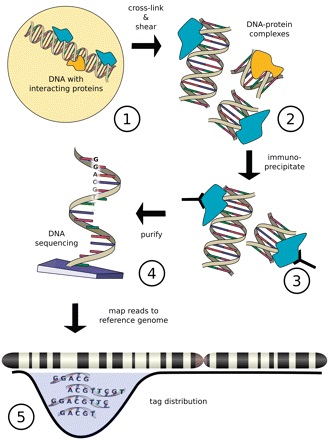
ChIP (chromatin immunoprecipitation) method selectively enriches DNA sequences bounded to specific proteins in vivo. The process begins with the fixation of chromatin with formaldehyde through cross-linking of the DNA binding proteins with DNA. Subsequently, shearing of the chromatin into smaller fragments (200-600 bp) by sonication, followed by immunoprecipitation of the DNA-protein complex using an antibody specific to the target proteins. In the next step, the crosslinks are reversed, DNA is isolated from the complexes and PCR (polymerase chain reaction) amplified. Finally, the DNA fragments are sequenced using NGS technology. The data or the reads generated from the sequencing run are then mapped to the genome and the protein binding DNA sites are identified (Figure 1). There are several sequencing platforms available that are suitable for ChIP-Seq assay, including, Illumina Genome Analyzer, Applied Biosystems SOLID, Roche 454-FLX and Helicos HeliScope, etc. However, the majority of the papers published so far have been analyzed on Illumina platforms. Many algorithms such as MACS (Model-based analysis of ChIP-seq), ChIPDiff and ODIN (One-stege Differential) are available for analyzing the protein-DNA interactions in the data generated by ChIP-Seq. This is referred to as peak detection which involves the detection of genomic regions where more reads are mapped than expected by chance.
Applications of ChIP-Seq
ChIP-Seq technique has been employed in many studies related to transcription factors, gene regulation to better understand diseases like breast and prostate cancer, nucleosome positioning, epigenetic regulation through histone modification.
Advantages of ChIP-Seq
ChIP-Seq has evolved as a potential technique in comparison to the previously used ChIP-chip assay.
- It depicts a higher base-pair resolution.
- It shows lesser artifacts and low noise generation.
- It exhibits a broad dynamic coverage and linear intensity signals.
- It depicts large genomic coverage.
- It requires very less input DNA as compared to ChIP-chip.
- It is compatible with various input DNA samples and can be utilized to analyze any species with an available sequenced genome.
Sensitivity of ChIP-Seq
Some limitations of the ChIP-Seq are as follows:
- This technique is more biased towards GC-rich content in fragment selection during the steps of library selection and amplification.
- Like any other technique, ChIP-Seq also produces artifacts or noise, although the improvements in the technology have considerably reduced the errors.
- Sequencing depth is an important limitation of this technology. The sequencing depth directly influences the cost of the run. If the requirement is to map the entire large genome with higher sensitivity, the cost automatically increases. However, the decreasing costs of the sequencing reactions and library preparation will eventually make ChIP-Seq a favorable method for various ChIP experiments.
References:
- Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods. 2007; 4(8):651-7.
- Park, P. J. ChIP-Seq: advantages and challenges of a maturing technology. Nature Genetics. 2009; 10:669-80.
- https://en.wikipedia.org/wiki/ChIP-sequencing.
- Precise analysis of DNA-protein binding sequences. https://sapac.illumina.com/techniques/sequencing/dna-sequencing/chip-seq.html.
- An introduction to ChIP-Seq. https://bitesizebio.com/13541/an-introduction-to-chip-seq/.
- Schmidt, D. et al. ChIP-Seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009; 48(3):240-48.