Genome sequencing technologies have come a long way since 2003 with the completion of the Human genome project. The first generation sequencing technology (FGS) famously known as Sanger’s sequencing provided a breakthrough in the field of genomics as it made possible the decoding of the complete genome sequence for the first time. However, they have several drawbacks: long run times, limited data throughput, higher cost and intensive labor. The second-generation sequencing (SGS) techniques have been a historic development over first-generation sequencing (FGS) technique by providing high-throughput data from multiple samples at a highly reduced cost and run time. However, they generate short read lengths (~few hundred bases) which is a major limitation in genome assembly, detection of highly complex genome regions, detection of gene isoforms and detection of epigenetic modifications known as methylation.
In order to overcome these challenges, Pac Biosciences (PacBio) has developed a novel technology, Single Molecule Real-Time (SMRT) Sequencing, generating longer reads (10 kb-60 kb) making them an excellent alternative to the unsolved problems. This technology utilizes the process of DNA replication and enables real-time observation of DNA synthesis, making it completely distinct from SGS and is termed as third-generation sequencing (TGS) technology. In 2011, the very first commercial platform based on PacBio SMRT sequencing technology was introduced PacBio RS System (using P1-C1 chemistry). The RS system generated longer read lengths (~2,500 bp to 23,000 bp) with a mean read length of 1,500 bp. Later with the improvement in the chemistry, a highly advanced and new version platform came to the market, PacBio RSII System (using P6-C4 chemistry) generating high-quality longer reads (~20 kb to 60 kb) with a mean read length of ~10kb. Recently, PacBio has commercially introduced a new platform Sequel system with the same technology and chemistry as PacBio RSII System but with higher throughput and decreased cost. It is capable of generating seven times more high-throughput reads per SMRT cell. The market cost of this system is half the amount of the previous platforms and claims to deliver a 10X human genome coverage in a single day.
SMRT sequencing mechanism
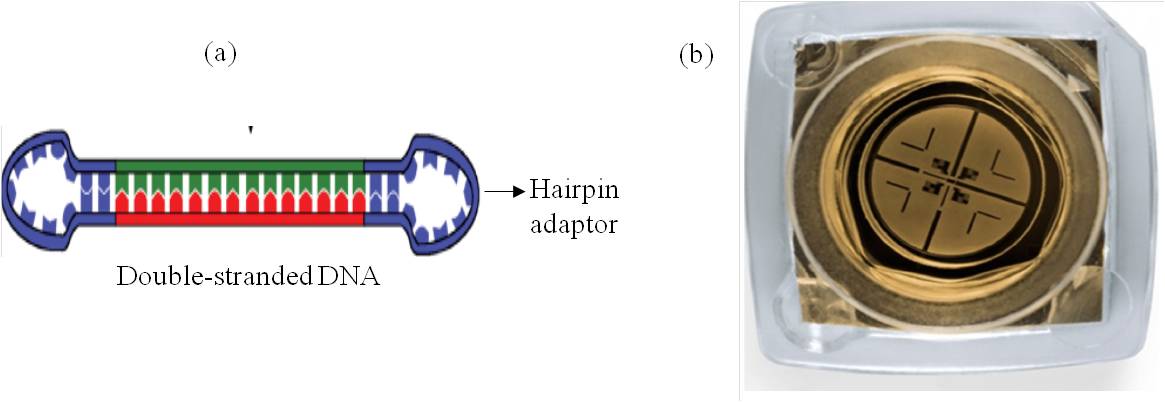
This technology collects sequence information as it enables real-time detection of nucleotide incorporation event during the replication process of the target DNA molecule. The first step is the library preparation from input double-stranded DNA molecule. The template DNA termed as SMRT bell is basically prepared by ligating hairpin adaptors onto both the ends of a double-stranded DNA molecule (Figure 1a). The sequencing is performed by loading SMRT bell onto a specialized chip known as SMRT cell (Figure 1b) which contains numerous units called as a zero-mode waveguide (ZMW) approximately 150,000, thus creating the smallest structural unit for the observation of only a single nucleotide being added by the DNA polymerase.
Image Credit: clpmag.com
At the bottom of each ZMW, there is a single immobilized polymerase which binds to either of the hairpin adaptors of the input SMRT bell and initiates the replication process (Figure 2a). The four nucleotides bases labeled with different fluorescent dyes are then added to the SMRT cell, and as each nucleotide is incorporated to the parental template a distinct fluorescent color is emitted which identifies the incorporated base. Once the nucleotide base is incorporated by the polymerase, the fluorescent dye is cleaved-off from the nucleotide and diffuses out of the detection unit (ZMW) finally ending the fluorescence signal (Figure 2b). The replication process takes place simultaneously in all the ZMW’s of the SMRT cell and is recorded by a “movie” of fluorescent pulses. The fluorescent pulses corresponding to each ZMW is then interpreted in the form of a sequence of bases known as continuous long read (CLR).
Strengths of PacBio SMRT sequencing
Recently we have witnessed a great rise in the use of SMRT sequencing owing to their various benefits:
- Longer read lengths– SMRT sequencing technology is capable of producing longest average read length (~30 kb) in comparison to other sequencing platforms.
- Single molecule resolution
- Uniform coverage- SMRT sequencing is highly efficient in sequencing the entire genomes with uniform coverage. This feature enables the sequencing through low-diversity and palindromic regions of the genome.
- Consensus accuracy- This sequencing technology provides exceptional sequencing data with an unmatched consensus accuracy (99.999%). This consensus accuracy, extremely low sequencing biases and precise mapping of the sequencing reads results in accurate detection of variants.
- Detection of epigenetic modifications- With this technique, researchers can easily detect DNA base modifications and very easily measure the DNA base incorporation rate during the sequencing process.
- Quick and efficient- This technology is very quick and highly efficient in generating data due to its easy and fast workflow.
Disadvantages of PacBio SMRT sequencing
SMRT sequencing technology also has some drawbacks:
- The throughput generated by SMRT sequencing is considerably low as compared to SGS techniques. The reason being, only 50%-60% of the total ZMWs produces successful sequencing reads, either due to no DNA template or failure in the proper binding of the DNA polymerase at the bottom of the ZMW.
- The other major drawback of SMRT sequencing is the error rate which is higher (11%-15%) in comparison to the SGS platforms. In order to generate >99.99% accurate data, it requires coverage of 15 times passage over, which greatly impacts the time and cost.
Applications of PacBio SMRT sequencing
In the past few years, PacBio SMRT sequencing has emerged as groundbreaking technology owing to its desirable attributes in the field of life science.
- De novo genome assembly – The most important application of SMRT sequencing is to generate complete genome assemblies of the novel genomes or species. Credit goes to the long reads produced by this platform which facilitates the assembly of high-quality and finished genomes. It is routinely being used for microbial genome assemblies.
- Transcript sequencing using Iso-Seq method – In higher eukaryotic organisms, almost all the genes exhibit alternative splicing leading to the generation of numerous isoforms. It is actually surprising to see that the alternatively spliced isoforms from a single gene exhibit entirely distinct functions. SMRT sequencing allows transcript sequencing up to 10 kb by following the Iso-Seq method. The Iso-seq method has achieved remarkable attention for its ability to characterize alternative splicing and explore full-length isoforms.
- Characterization of structural variations – SMRT sequencing enables characterization of complex genome regions and makes it easier for resolving complex structural issues such as SNPs, indels, structure variants and haplotypes. The long reads produced by this platform are capable for variant calling as it determines the precise location, allelic sequence, and size in comparison to the short reads.
- Epigenetic methylation studies – DNA base modifications are known as DNA methylation are key regulators for a number of biological processes such as gene expression, host-pathogen interaction, gene silencing, DNA damage and repair. The PacBio SMRT sequencing monitors the single nucleotide base addition by measuring the kinetic properties of the base incorporation to the template strand during the sequencing process. This kinetic property enables direct detection of various base modifications.
References:
- SMRT Sequencing. Pacific Biosciences. https://www.pacb.com/smrt-science/smrt-sequencing/
- The Advantages of SMRT Sequencing. Genome Biology. 2013, 14:405.
- PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics. 2015,13: 278–289.
- Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acid Research. 2018, 46:2159-2168.